The Flagpole
MHC (major histocompatibility complex), also known as HLA (human leukocyte antigen) is the entire set of genes that code for proteins involved in antigen presentation. Very simply, you could think of HLA as analogous to a “flagpole waving a flag” to notify the immune system of a particular antigen. Hey Immune System, is this “self” or “non-self?” or in other words, “Are you foreign???” HLA has also been described as a billboard advertising peptides to the mighty CD8+ and CD4+ T cells as they provide immune surveillance.
MHC Class I and Class II have distinct roles and a few differences include the following:
Binding Groove The peptide-binding groove or binding pocket where the antigen sits is unique between Class I and Class II. It has been established that in order for a T-cell to be alerted, the peptide has to be stably bound within this cleft. Hence, peptide-binding affinity is a critical factor in putative neoantigen candidate criteria. For MHC class I, the peptides are ~8 to 11 amino acids in size and certain positions within the pocket anchor the peptide. In Class I antigen presentation, position 2 and position 9 are thought to be the essential anchor residues. The peptide can lay flat in the MHC Class I binding pocket or can “bulge out”. For Class II, instead of having a peptide that is constrained within this cleft, especially at the carboxyl terminus, class II peptides can be much larger in size (10 to >30 amino acids). Peptides presented on HLA Class II can hang off the side of the groove, much like how a hotdog hangs over the edge of a bun.
T cell Interactions In general, CD8+ T cells, or cytotoxic T cells, recognize tumor antigens presented on HLA Class I, while CD4+ T cells, or T helper cells, recognize those epitopes presented on HLA Class II.
Antigen Processing How do these peptide fragments or antigens make their way to the cell surface to be displayed for immunosurveillance? There are a variety of processing steps that must occur in order to properly present epitopes to T-cells. This involves a complex series of compartments, chaperones, and enzymes for both class I and class II. In brief for Class I, 1) the proteosome degrades the peptide into fragments, 2) the antigen is transported into the Endoplasmic Reticulum (ER) by TAP (antigen transporter), 3) chapperoned into the Golgi and then 4) bound peptides are presented externally to CD+8 T-cells. While for Class II, 1) exogenous antigens are broken down by proteases, 2) the MHC heterodimers assemble a pseudopeptide in the ER, 3) antigens are guided out of the ER and thru the Golgi, 4) the pseudo peptide is degraded to a CLIP peptide, 5) catalyst HLA-DM removes CLIP and adds the antigen to the class II molecule, and then 6) is exported to the surface to interact with CD4+ T-helper cells. In this pathway, there are many opportunities for mutations to impact pathway function. As evidenced in the literature, defective antigen presentation by HLA class I has been shown as a mechanism of cancer immune escape (Rodríguez 2017). Loss of HLA class I or beta 2 microglobulin (B2M), an essential component of the HLA class I molecule, can inhibit the function of HLA to recruit immune cells within tumors. Therefore without intact antigen presentation, this leads to immunotherapy resistance.
NGS-based HLA typing
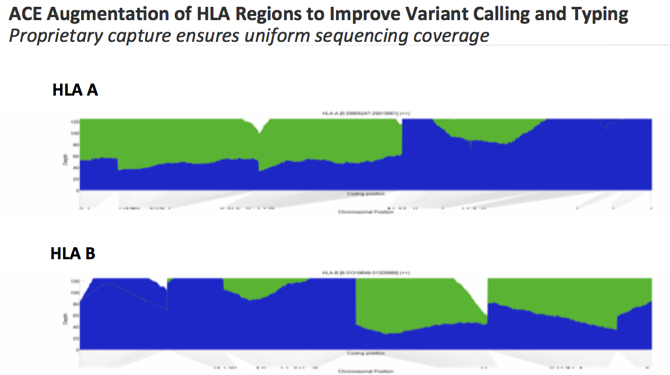
HLA typing for organ transplantation or tissue matching purposes has typically been performed using PCR-based methodology. For this application, sequence specific primers are designed against the HLA loci in order to evaluate compatibility between donor and recipient. As the HLA genetic loci are very polymorphic in nature, it has been thought to be more challenging to perform HLA typing from NGS data. However, within the Personalis proprietary ACE design, we’ve optimized and augmented the coverage of the HLA genes in our exome and transcriptome. As shown in the diagram below for the HLA A and HLA B loci, blue shows the sequencing coverage using a standard exome assay design. The green shows the enhanced coverage provided with the proprietary ACE capture. Thus, “blue plus green” displays the uniform exome sequencing quality from the augmented ACE exome to enable accurate variant calling for HLA typing.
To investigate HLA typing performance using the ACE Exome approach, we sought to validate hundreds of HLA alleles across both MHC class I and II. We procured samples from the International Histocompatibility Working Group (IHWG) that had results from orthogonal clinical grade HLA Testing (PCR-based). Blinded samples from a collaborator provided further samples to include in the study. Upon comparing NGS-based to clinical HLA typing, we found very high concordance between the results from both methods. This analysis was performed using the patient blood sample and 4-digit HLA characterization. Overall, for both Class I and select class II alleles, final results showed a 96% concordance. This data demonstrates the ACE exome approach as robust and accurate for HLA identification.
HLA as a module for neoantigen identification
As mentioned above, the ability for the predicted neoantigen to have a strong binding affinity within the pocket provides evidence for the likelihood that it will be appropriately presented on the MHC. In silico prediction algorithms determine the affinity for the peptide to form a stable complex on the MHC. These software programs can predict the binding affinity of both class I and class II neoepitopes. Currently, the accuracy of these programs has been questioned, yet this remains a critical first step in determining prospective neoantigen targets. In the first-in-human clinical trials using personalized neoantigen based vaccines, HLA binding affinity was a key criterion for helping determine candidates (Ott et al. 2017, Sahin et al. 2017).
HLA as a Biomarker
Can a patient’s HLA type provide clues to how they may respond to checkpoint inhibitor therapy? Investigators at Memorial Sloan Kettering recently published results from a study determining if zygosity at HLA loci or if certain HLA alleles may influence survival to anti-CTLA-4 or anti-PD-1/PD-L1 therapy (Chowell et al. 2017). Their hypothesis was that greater diversity within the HLA I molecule would result in a larger repertoire of neoantigens to be presented. Using HLA genotype data from >1,500 cancer patients, homozygosity in at least one HLA class I locus was associated with reduced survival for checkpoint inhibitor therapy. Further, somatic alterations in HLA were likewise shown to be indicative of patient response. The authors also analyzed tumor specimen for Loss of Heterozygosity (LOH) and patients with LOH at HLA class I loci were also associated with reduced survival following checkpoint blockade immunotherapy.
Understanding patient HLA genotype can be utilized in both personalized cancer vaccine development as well as in the immunotherapy biomarker discovery. Thus, as the clinical utility and impact continues to mount, having accurate HLA typing results becomes more essential than ever before.