Q&A with John West, Founding CEO of Personalis
When Personalis was founded in 2011, technological advances had just brought whole genome sequencing into reach, opening the possibility to improve patient outcomes. This is the goal that Personalis was founded on, and it’s the goal that still guides us to this day.
In just over a decade, Personalis has led the industry by sequencing over 150,000 human genomes, the most of any for-profit company in the United States. Among these genomes, thousands are derived from tiny, biochemically complex samples, collected from patients with cancer. Working with these samples is far from trivial, but doing it on such a scale in a CLIA/CAP laboratory is something else entirely. It has required a relentless commitment to quality, implementation of advanced automation systems to reduce human error, the development of data storage systems capable of holding tens of petabytes of data, and cutting-edge informatics that enable everything from sample tracking to variant calling and clinical reporting.
Personalis has been building and innovating for over eleven years now, but we believe we’ve only just begun. In this multipart series, we’ll be talking to Personalis’ founder and CEO, John West, about this journey and where we’re headed, both as a company and industry.
Q: We’ve sometimes said that Personalis is the nexus point for many pioneering efforts in the genomics space. Why is that?
A: Personalis has always been a place that attracts expertise, I think because we are all motivated to look beyond today’s technology, seeking better outcomes for patients. That’s very attractive to people who have built their careers at the intersection of genomic technologies and medicine.
I’m no different. I began work on DNA sequencing automation, with a focus on the human genome, nearly forty years ago. Over my career, I’ve been really able to see and participate in the evolution of whole genome sequencing.
In 1988, my first system was commercialized and could generate six thousand bases of DNA sequencing data in a single run. By fourteen years later, I was managing the DNA sequencing business at Applied Biosystems, and in 2002 my team introduced the ABI 3730xl capillary sequencer, which could generate six million bases in a single run. In 2004, I became CEO of Solexa and within four years, we had developed the technology used to sequence the first cancer genome. In 2007, I sold Solexa to Illumina and became their first senior vice president of DNA sequencing.
Illumina’s Genome Analyzer IIx system could generate six billion bases in a single run. That was the foundation for the Illumina DNA sequencing technology we know today. And you can see how it’s a straight throughline to Personalis today. In our lab, we use the Illumina NovaSeq systems which can now generate six trillion bases in a single run. Already in my career, I’ve seen a billion-fold increase in the scale of sequencing technology.
It’s natural for us at Personalis, then, to plan for continued advances. We know the $100 genome is coming – Illumina first hinted at it when they introduced the NovaSeq system in 2017. We know that others have the same goal, too, such as Ultima Genomics. Personalis has known the Ultima team since it was founded and took delivery of one of their first sequencing systems this past January.
I mention this because it speaks to our team’s experience: we’ve helped drive the advance of sequencing technology, giving us insight in how best to use it and what to build on top of it.
The history of the human genome has involved numerous companies, and our team has experience from many of them. Christian Haudenschild, our VP of Operations, and Laurie Goodman, who leads our Medical Affairs team, worked at Lynx Therapeutics, which developed the first commercial massively parallel sequencing system, MPSS, back in the 1990’s.
John Lyle, our VP of Assay Development, and Steve Moore, our General Counsel, each worked at PacBio for a decade, driving the development of long read DNA sequencing technologies that eventually led to the first telomere-to-telomere human genome. And in the 1990’s, Rich Chen, our Chief Medical Officer and SVP of R&D, co-founded Ingenuity Systems, one of the first informatics companies to leverage genome-scale gene expression data and advance the understanding of complex gene-interaction pathways. Finally, Massimo Morra, our VP of QA & Compliance, helped build Correlagen, one of the first diagnostic companies based on DNA sequencing.
All of this is to say, we have a very deep bench of experience.
Q: How had Personalis, at its founding, come to view this technology as one that could be transformative in the clinic?
A: Although Personalis is pioneering many different technologies, building on our genome sequencing expertise, from our inception we’ve been committed to medical applications.
In 2009, when Illumina first began to offer personal genome sequencing as a service, I no longer worked at the company, but jumped at the chance and signed up. My family, all of us healthy, were the first named family of four to be sequenced by Illumina. This was important because family sequencing can be used to phase the individual genomes, resolving what is called compound-heterozygosity. Individual genes can contain more than one variant, and the medical impact of these variants can be very different if they impact both copies of a chromosome versus just one. We wanted to pioneer this at a whole genome level.
While we were at the forefront of healthy family genome sequencing, Euan Ashley was leading a group at Stanford on a project to use whole human genome sequencing in a serious medical case. Their work, published in 2010, showed that a human genome sequence can be an important tool for advising an individual patient. When I spoke with the Stanford group, it became clear that we were interested in the same goals and clinical research questions, so we decided to team up.
As is now described in Euan’s book, The Genome Odyssey: Medical Mysteries and the Incredible Quest to Solve Them, this partnership led to the founding of Personalis. By September 2011, Personalis was incorporated, had arranged its $20M Series-A venture financing, and had published its first journal article extending some of the Stanford team’s methods to my family’s phased genome data.
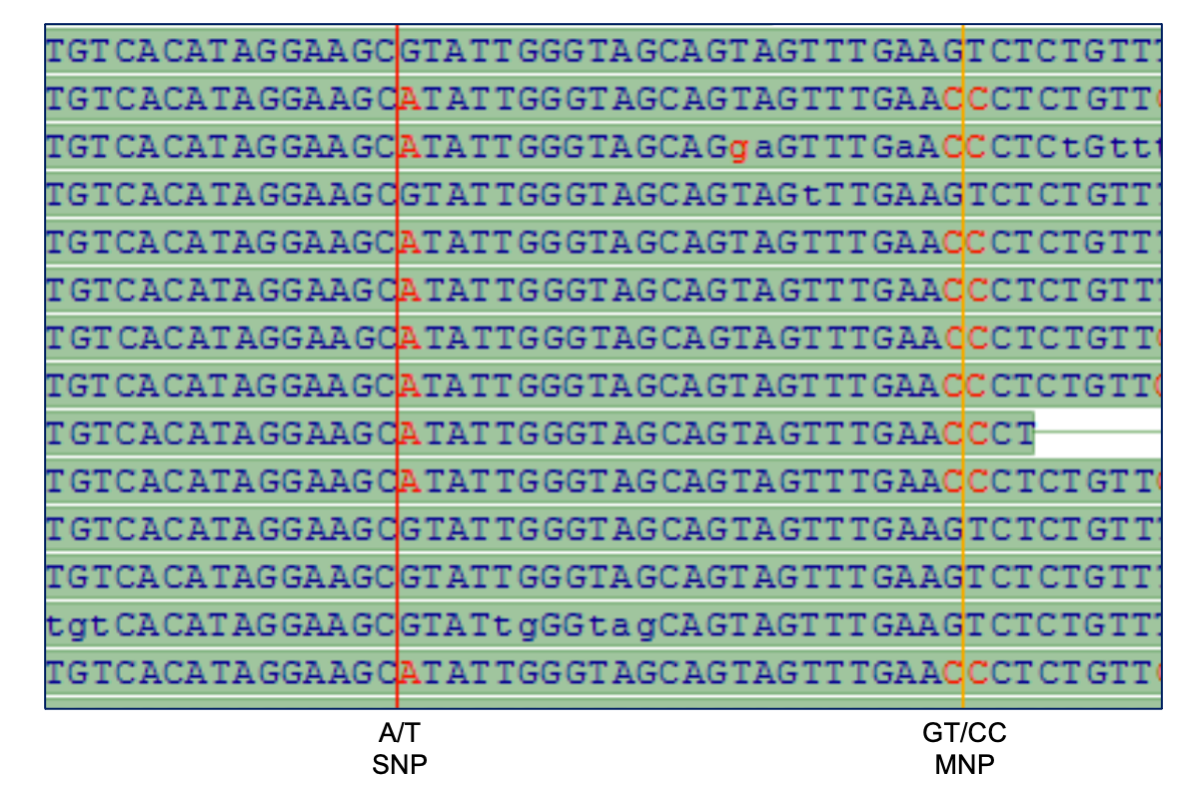
Figure 1: Determining a local haplotype by phasing a SNP with an MNP, by detecting them both on the same sequence reads. Data we presented at the AGBT conference, February 2011. We subsequently went beyond this, phasing entire genes using genome data from a family of four.
You can see that, even before Personalis was founded, the clinical application of genome-scale sequencing was squarely in our sights.
Q: Personalis has been focused on cancer for many years now. How does the company leverage its deep experience with whole genome sequencing, in cancer ?
A: When Personalis was founded, a whole genome was considered the ultimate and complete data set, comprehensively covering all of a person’s genetics. A genome was something to be sequenced once in a person’s lifetime. In cancer however, nothing could be further from the truth. There is enormous additional information in the RNA, in the immune repertoire, and in the viral microbiome. The situation is also very dynamic, with tumors growing exponentially and metastasizing against an onslaught of surgical and therapeutic interventions. Experience with large scale human genome sequencing has prepared us fantastically for that. We have one of the largest sequencing labs in the world, and can comfortably plan to sequence a trillion bases per patient as it is needed.

Figure 2: Large scale human genome sequencing requires state of the art DNA sequencing systems, but also sophisticated laboratory robotics and data systems. Left: Our first six Illumina NovaSeq instruments, in 2018. Right: Our first Ultima Genomics instrument, January 2022. Lower: Some of our more than 20 advanced laboratory pipetting robot systems.
Whole genome work also brings an informatics and clinical mindset of comprehensiveness, in an environment of great complexity. This has held us in good stead as we have pioneered in cancer. All of our cancer products, whether whole genome, exome, or transcriptome, encompass the compete set of ~20,000 genes. When others developed products for tracking molecular residual disease (MRD) based on just a few percent of somatic variants, we saw the limitations of that approach and built our first product with a whole genome front end and capacity to follow thousands of variants. This is not data for the sake of data, or for academic scientific research. Clinicians need sensitive tests without clinically actionable blind spots.
The field of whole genome sequencing also has an ethos of ultimately being for all patients. Personalis has taken this into our cancer business with pan-cancer offerings. We have pioneered the use of whole genome sequencing at the front end of our MRD assay, with this in mind. The largest segments of the cancer survivor patient population are those originally diagnosed with breast and prostate cancer. Many of these patients have very low mutational burden, low shedding, and surgical resection early, when the residual disease may also be very low. They may not have the convenient, perhaps more easily detectable, mutational signatures of other cancer types. Still, we need tests which will work for them as well. As DNA sequencing technologies become more and more accurate, we will want to leverage all of the somatic variants available, to capture as much signal as possible in these challenging patient populations. Using whole genome sequencing gives us access to thousands of somatic variants, in almost every tumor.
We also need to accept the reality of very limited sample amounts. Because of this, we plan to validate our MRD tests down to 5 nanograms of input. By leveraging almost two thousand positions across the genome, our MRD test is designed to stay sensitive, even at these low levels. As in the whole genome sequencing world, our goal is to support all patients.
As you can see from this short conversation with John, Personalis has a deep history in genomics that predates its founding. This experience enabled the Personalis team to hit the ground running as soon as it was formed. It’s also clear that from its inception, Personalis was focused on building tomorrow’s gold standard, ultimately to improve patient outcomes with more insightful diagnostics and faster pharmaceutical development. Subsequent blogs in this series will explore this ethos and what John sees on the horizon for Personalis and the genomics industry as a whole.

Figure 3: Personalis company celebration, on completion of our 100,000th whole genome sequence, Jan 15, 2021, by Zoom.