In a study led by University of Pennsylvania and the Parker Institute for Cancer Immunotherapy, Personalis is a co-author on a poster being presented at the American Society of Clinical Oncology (ASCO) Annual Meeting 2022, titled, “Circulating KRAS variant-specific shedding and association with survival in patients with metastatic pancreatic ductal adenocarcinoma (mPDAC) receiving chemoimmunotherapy.”
As background, circulating tumor DNA (ctDNA) is increasingly used as a prognostic marker, with high ctDNA shedding associated with poor survival for patients. Researchers have reported gene-, but not variant-specific, differences in ctDNA shedding, while tumor burden, mitotic rate, and cell death rate have been proposed as contributors.
The study authors investigated associations of ctDNA shedding for the two most common metastatic pancreatic ductal adenocarcinoma (mPDAC) KRAS variants, G12D and G12V, with tumor burden, mitotic score, and overall survival. The study concludes: “As clinical ctDNA tests become more widely used, further investigation of variant-specific shedding may be key for proper interpretation of ctDNA tests.”
In this Q&A with Personalis co-author Lee McDaniel, MD, Senior Bioinformatics Scientist, he provides his own insights into the study and its findings.
Q: What did we hope to learn by understanding variant-specific shedding? Why does shedding matter?
A: G12D and G12V are fascinating variants because they reside in an essential portion of the KRAS protein. If KRAS is a skeleton key that unlocks the doors to multiple downstream pathways, G12D and G12V are little changes to the shape of the key – changes that now unlock different pathway doors in each case. We’re still trying to figure out why activating different pathways yields different shedding characteristics. It could be different gradations of cellular turnover and apoptosis, different rates of tumor growth, or the seeding of anatomic sites with greater or lesser propensities for shedding.
Understanding the genomics of shedding gives meaning to interpreting a scenario where we have clearance, where the shedding has stopped: is a high-shedding tumor eradicated, or is this a low-shedding tumor where we couldn’t detect a vanishingly small signal? The latter case demonstrates a need for maximally sensitive detection of ctDNA.
Q: Why is this topic important to the field?
A: Our collaborative group has discovered that clearance of G12D shedding is predictive of survival, but not G12V. Survival prediction is important because it may aid clinical decision-making by increasing therapeutic intensity to improve outcomes. This study suggests that specific amino acid substitutions should be considered because that prediction only seems to apply in certain cases. In two different cohorts, we demonstrated that G12D shedding at baseline is predictive of overall survival, which opens the door for more intensive therapy for such risk-stratified patients in the future. the study shows that even a single substitution difference at 1 in 3 billion base pairs in our genome needs to be considered when conducting these predictive approaches.
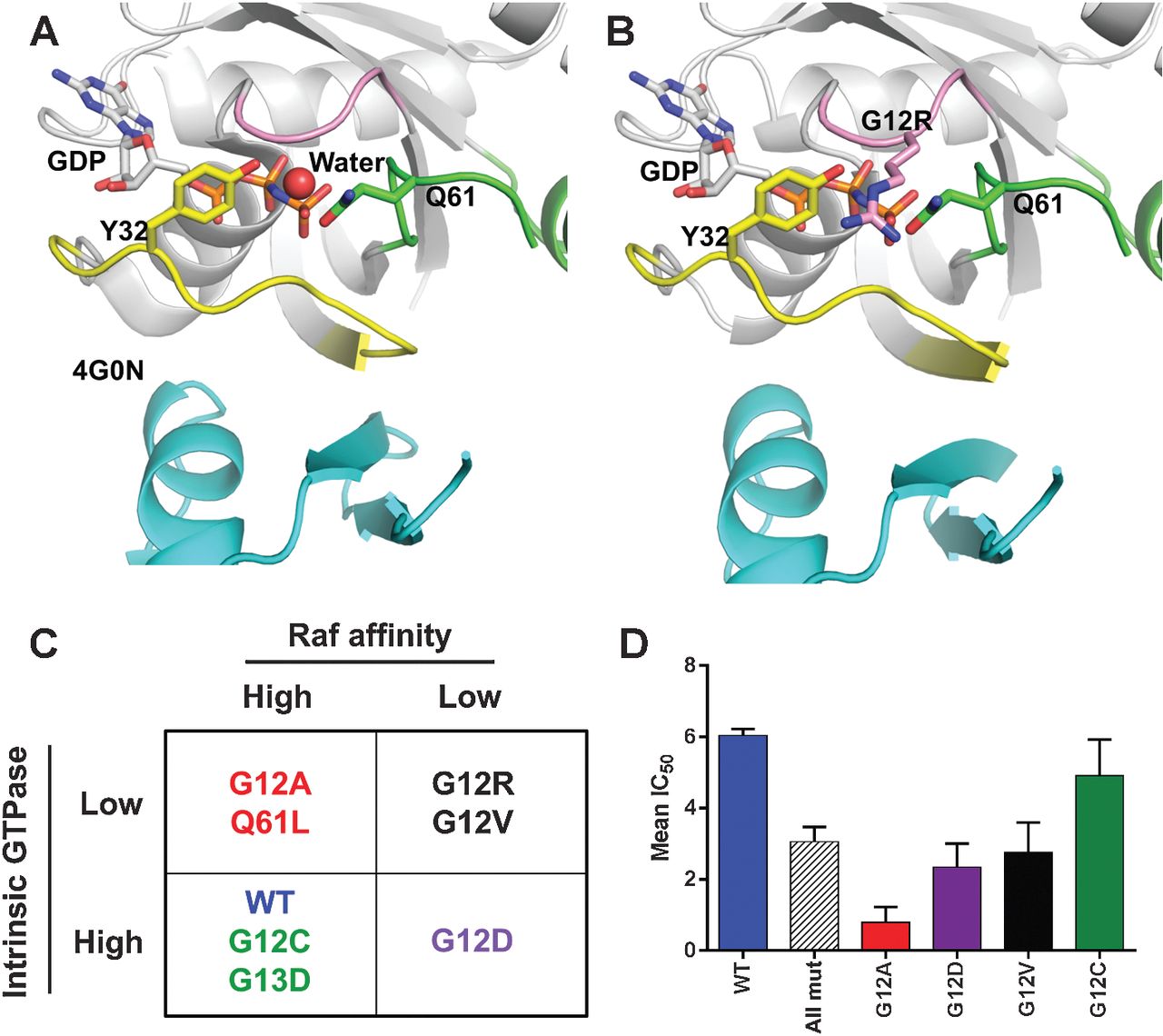
Figure 1: Predicted impact of KRAS mutations on RAF kinase signaling.
A, published structure of HRAS (white) in complex with the RBD of RAF kinase (cyan, PDB ID: 4G0N). In the complex switch I (yellow) and switch II (green) are in the closed conformation and Y32 and Q61 rotate toward the γ phosphate of GMPPNP. B, model of G12R KRAS bound to the RBD of RAF kinase prepared by aligning the G12R-mutant structure to A. In this model, the P-loop (pink) R12 side chain atoms clash with both Q61 and Y32 residues and displaces a coordinated solvent molecule. These predicted perturbations may lead to some destabilization of both switch I (yellow) and switch II (green) regions and a perhaps explain the decrease in affinity observed for G12R KRAS. C, proposed classification scheme of KRAS mutants based on intrinsic GTP hydrolysis rates and relative affinity for RAF kinase. D, KRAS-mutant cell lines show differential sensitivity to the MEK inhibitor PD-0325901. Publicly available sequencing and pharmacologic data from the cancer cell line encyclopedia were downloaded from the Broad institute and mean IC50 values for each KRAS mutant calculated ±SE.
Q: How do you see shedding impacting our understanding of applications such as tumor progression or treatment resistance?
A: Shedding is consequent to three factors. First, the size of the tumor: how many tumor cells are there that might shed? Second, where is the tumor? Different anatomic sites and tissues have vastly different degrees of vascularization. Third, what biomolecular characteristics in these tumor cells cause them to either shed their DNA into the body, versus degrade it and not shed?
With an accurate and precise shedding model, we can make inferences about tumor biology or tumor size. For example, if we measure shedding and estimate per-cell shedding based on variant and RNA expression characteristics, we can estimate the physical tumor size. Conversely, if we have a measurement of tumor size and a measurement of shedding, we might be able to say something about what’s happening on a cellular level. Such findings could be directly relevant to clinical care: to estimate whole-body tumor volume without imaging (for tumor progression) and to assess response to therapy (for treatment resistance).
In this poster we demonstrated that looking just at a single position in the genome, a single nucleotide translated into an amino acid, we were able to predict overall survival based on baseline shedding. Already demonstrated in two cohorts, with further investigation this may serve to predict therapeutic response. In this cohort, shedding was undetectable for a significant portion of patients, which is unsurprising since we’re trying to detect just a tiny drop of DNA in an ocean of DNA fragments.
Beyond this study, that’s one of the reasons why Personalis is excited about NeXT Personal™, which expands the search to thousands of sites – we’re hoping to detect ctDNA fragments when none were detected before by virtue of testing so many sites. It’s the difference between taking a single light measurement with one pixel, and taking a photograph with thousands of pixels. We’re hoping that larger picture will uncover new associations with therapeutic response.
Q: How was ImmunoID NeXT™ integrated with droplet digital PCR to explore shedding in this study?
A: Using ImmunoID NeXT’s DNA-seq and RNA-seq capabilities, as well as droplet digital PCR (ddPCR), we were able to confirm KRAS genotypes with these three different nucleic acid genotyping methods. More importantly, the task of understanding shedding biology relied on three measurements: ddPCR, ImmunoID NeXT, and RECIST measurements of tumor size. Our group has sought to leverage RNA-seq data to build shedding related models, and to do this we must adjust for tumor volume. All else being equal, a physically large tumor should shed more than a tiny tumor. We can adjust the shedding measurement by the size and location of tumors, yielding a normalized measurement for our ImmunoID NeXT-based cellular models.
We took advantage of the robust RNA-seq features from ImmunoID NeXT while investigating the hypothesis leading to this poster. One of our hypotheses was that mitotic rate might be related to shedding, informed by prior publications using mitotic rate as part of a theoretical model. The reasoning behind this is the idea that increased mitosis is matched with increased apoptosis or necrosis, that there is differential “churn” of cells in some tumors. By layering mitotic score calculations on top of ImmunoID NeXT RNA-seq data, we found that mitotic rate was predictive of shedding in G12D but not G12V tumors. RNA-seq supported this study in several other ways: PDAC subtyping and GSEA analysis. Using GSEA, we are able to identify pathways associated with differential shedding to understand why we see this differential association.
Q: Can the findings be applied to other cancer types? If so, which ones?
A: The KRAS G12 mutation site plays a prominent role in several other common cancers such as colorectal cancer and NSCLC, including a KRAS mutation not commonly seen in PDAC, G12C. It would be straightforward to conduct a parallel study in these other cancers that commonly have KRAS mutations.
On a more general level, this study introduces new approaches to link together shedding, tumor size, and genomics. While in this study we only probed the KRAS G12 locus for shedding measurement, we’re going to see new studies where hundreds and thousands of genomic loci are evaluated for shedding in many cancer types. Our study has developed the sorts of analytic methods that can be extended to these new state-of- the-art panels. At Personalis, using NeXT Personal, we’re also hoping to not only look at KRAS alleles in CRC and NSCLC, but also to expand the view into other prominent oncogenes and how they might drive shedding.
Contact us to learn more about partnerships.
All products described here are for Research Use Only and not for use in diagnostic procedures (except as specifically noted).